Linear Discriminant Analysis (LDA)
In this example, we will consider the application of linear discriminant analysis (LDA) to the "Irises of Fischer" dataset.A comparison with the principal component analysis (PCA) will also be conducted.
Linear discriminant analysis (LDA) is a statistical analysis method that allows you to find a linear combination of features to divide observations into two classes.
Pkg.add(["MultivariateStats", "RDatasets"])
Let's assume that the samples of positive and negative classes have average values:
(for the positive class),
(for the negative class),
as well as covariance matrices and .
According to the Fisher criterion for linear discriminant, the optimal projection direction is given by the formula:
where — an arbitrary non-negative coefficient.
Installing and connecting the necessary libraries:
using MultivariateStats, RDatasets
Downloading data from Fischer's Irises dataset:
iris = dataset("datasets", "iris")
Extracting a matrix of observational objects with features from a dataset - X and the class vectors of these objects - X_labels:
X = Matrix(iris[1:2:end,1:4])'
X_labels = Vector(iris[1:2:end,5])
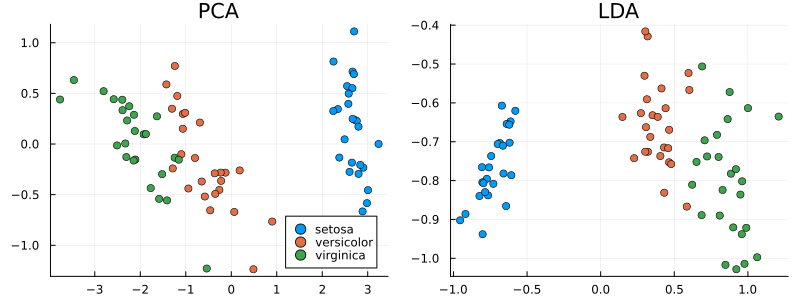
Let's compare linear discriminant analysis with the PCA (principal component method) method.
Learning the PCA model:
pca = fit(PCA, X; maxoutdim=2)
Applying PCA to data:
Ypca = predict(pca, X)
Learning the LDA model:
lda = fit(MulticlassLDA, X, X_labels; outdim=2);
Applying LDA to data:
Ylda = predict(lda, X)
Visualization of results:
using Plots
p = plot(layout=(1,2), size=(800,300))
for s in ["setosa", "versicolor", "virginica"]
points = Ypca[:,X_labels.==s]
scatter!(p[1], points[1,:],points[2,:], label=s)
points = Ylda[:,X_labels.==s]
scatter!(p[2], points[1,:],points[2,:], label=false, legend=:bottomleft)
end
plot!(p[1], title="PCA")
plot!(p[2], title="LDA")
Conclusions:
PCA and LDA are dimensionality reduction methods with different goals: PCA maximizes global data variance and is suitable for visualization without class labels, while LDA optimizes class separation using label information, which makes it effective for classification tasks. In the example with Fischer's Irises, LDA provided a clear separation of classes in the projection, while PCA retained the general data structure, but with overlapping classes. The choice of method depends on the task: PCA — for data analysis, LDA — for improving classification in the presence of marked classes.