Classification of LPI radars
Introduction
LPI radar (Low Probability of Intercept) systems are designed to evade detection by using special modulation and transmission schemes over wide frequency ranges with low power levels. In modern LPI radars, these conditions are usually achieved by transmitting a continuous wave (CW) signal. Detection and classification of LPI signals is a serious task. Recent advances show that deep learning techniques can be successfully applied to classify radar signals.
This example shows how to classify LPI radar signals using a neural network. Before submitting data to the ANN input, they undergo processing - we obtain data in the time-frequency domain from the complex signal, after which, using image processing, we obtain a binary spectrogram and a feature vector obtained using HoG.
Read more about data preprocessing below
The neural network itself consists of two branches. The first branch is a two-dimensional convolutional network with a binary spectrogram input. The second branch of the neural network is a one-dimensional convolutional network, the input of which is a vector of features extracted using HoG from a binary spectrogram. The feature data at the output of the two branches is combined and fed into the head of the INS - 2 fully connected layers that produce logits. And then, using cross-entropy, the predicted class labels for the signal are calculated.
Preparation for work
Importing the necessary packages
include("installPackages.jl");
using Pkg
Pkg.instantiate()
using SignalAnalysis
using DSP
using MAT
using Plots
using Images, ColorSchemes, Random
using ImageBinarization, ImageFiltering, ImageMorphology, ImageFeatures
using Flux, CUDA, cuDNN
using OneHotArrays
using StatisticalMeasures
include("show_sample.jl");
include("utils.jl");
CM = StatisticalMeasures.ConfusionMatrices;
Let's set the parameters that will be used in this work.
n = 64
hop=48
Fs = 20e6
nfft = 256;
threshold = 210 / 255
datadir = "data";
n_classes = 11;
lr = 1e-3
CLASSES = 1:n_classes;
Data overview
We will perform preprocessing for one selected signal.
First, we will download and read the signal from the file. data - a signal in a complex form
testpath = joinpath(datadir, "Frank")
files = readdir(testpath)
n_files = length(files)
ind = Random.rand(1:n_files)
filepath = joinpath(testpath, files[ind])
data = matread(filepath)["x"][:, 1];
Function show_sample calculates the spectrogram on the signal using the parameters for the STFT that we set above
z, fig = show_sample(data, n, hop, Fs, nfft);
fig
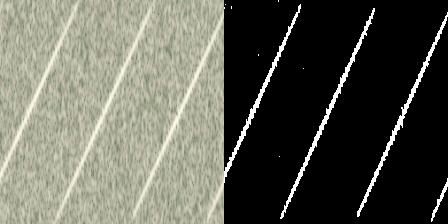
The getImage function allows you to obtain an RGB image from the calculated spectrogram. Well, getBinary calculates the binary image of the received spectrogram with a given threshold. The results are visualized below
img = getImage(z)
bnr_img = getBinary(img, threshold)
[img bnr_img]
Next, we will submit the resulting binary spectrogram to HoG for feature extraction. HoG calculates a histogram of gradient orientations (directions of brightness changes). At the output of the method, we obtain a vector of features that can be applied to the input of a one-dimensional convolutional network.
In general, this vector can be visualized. High columns mean that the border in the corresponding cell is strongly marked at a given angle; zero or almost zero values mean that there are no gradients in this direction in the cell.
featureHoG = calcHoG(bnr_img)
plot(featureHoG, legend=false, xticks=false, yticks=false)
Next, we will create a dataset structure that will iteratively perform all the pipeline done above for each signal. In addition, we will divide all the data into three sets - training, validation and test in standard proportions. We will also create data loaders with a batch size of 64.
subfolders = readdir(datadir)
classes = subfolders
paths = String[]
labels = Int[]
for (cls_idx, sub) in enumerate(classes)
folder = joinpath(datadir, sub)
for fname in readdir(folder)
push!(paths, joinpath(folder, fname))
push!(labels, cls_idx)
end
end
N = length(paths)
perm = randperm(N)
paths = paths[perm]
labels = labels[perm]
N = length(paths)
train_len = floor(Int, 0.7 * N)
val_len = floor(Int, 0.15 * N)
test_len = N - train_len - val_len
train_idx = 1:train_len
val_idx = (train_len+1):(train_len+val_len)
test_idx = (train_len+val_len+1):N
train_paths, train_labels = paths[train_idx], labels[train_idx]
val_paths, val_labels = paths[val_idx], labels[val_idx]
test_paths, test_labels = paths[test_idx], labels[test_idx]
struct LazyRadarDataseT
paths::Vector{String}
labels::Vector{Int}
n; hop; Fs; nfft; threshold
end
Base.getindex(ds::LazyRadarDataseT, i::Int) = begin
data = matread(ds.paths[i])["x"][:,1]
b, f = prepareSample(data, ds.n, ds.hop, ds.Fs, ds.nfft, ds.threshold)
return Float16.(b), Float16.(f), ds.labels[i]
end
function Base.getindex(ds::LazyRadarDataseT, idxs::AbstractVector{Int})
B = length(idxs)
b0, f0, _ = ds[idxs[1]]
nb, Tb = size(b0)
Tf = length(f0)
batch_b = Array{Float16}(undef, nb, Tb, 1, B)
batch_f = Array{Float16}(undef, Tf, 1, B)
batch_labels = Vector{Int}(undef, B)
for (j, i) in enumerate(idxs)
bj, fj, lbl = ds[i]
batch_b[:, :, :, j] = bj
batch_f[:,:, j] = fj
batch_labels[j] = lbl
end
return batch_b, batch_f, batch_labels
end
Base.length(ds::LazyRadarDataseT) = length(ds.paths)
train_ds = LazyRadarDataseT(train_paths, train_labels, n, hop, Fs, nfft, threshold)
val_ds = LazyRadarDataseT(val_paths, val_labels, n, hop, Fs, nfft, threshold)
test_ds = LazyRadarDataseT(test_paths, test_labels, n, hop, Fs, nfft, threshold);
# --- 4) We wrap them in a DataLoader — Flux will pull the necessary elements on the fly by index
batch_size = 128
train_loader = Flux.DataLoader(train_ds;
batchsize = batch_size,
shuffle = true,
partial = true,
parallel = true,
)
val_loader = Flux.DataLoader(val_ds; batchsize=batch_size, shuffle=true)
test_loader = Flux.DataLoader(test_ds; batchsize=batch_size, shuffle=true)
Let's define the device on which the network training will be performed.
device = CUDA.functional() ? gpu : cpu
Let's define the architecture of a neural network consisting of two branches and a head.
function makeBranch2dCnn(; p = 0.5)
Chain(
Conv((5,5), 1=>64, pad=1), relu,
x -> maxpool(x, (2,2)),
Conv((5,5), 64=>128, pad=1), relu,
x -> maxpool(x, (2,2)),
Conv((5,5), 128=>128, pad=1), relu,
x -> maxpool(x, (2,2)),
Conv((5,5), 128=>256, pad=1), relu,
x -> maxpool(x, (2,2)),
Conv((5,5), 256=>512, pad=1), relu,
Dropout(p),
x -> maxpool(x, (2,2)),
)
end
function makeBranch1dCnn(; p = 0.25)
Chain(
Conv((5,), 1=>8, pad=1), relu,
x -> maxpool(x, (2,)),
Conv((5,), 8=>64, pad=1), relu,
x -> maxpool(x, (2,)),
)
end
function makeHead()
head = Chain(
t -> begin
x1, x2 = t
v1 = reshape(x2, :, size(x2, 3))
v2 = reshape(x1, :, size(x1, 4))
return vcat(v1, v2)
end,
Dense(432576, 128, relu),
Dense(128, n_classes)
);
end
struct Net
branch1::Chain
branch2::Chain
head::Chain
end
function Net()
cnn2d = makeBranch2dCnn();
cnn1d = makeBranch1dCnn();
head = makeHead()
Net(cnn2d, cnn1d, head)
end
function (m::Net)(imgs, feat)
out2d = m.branch1(imgs)
out1d = m.branch2(feat)
return m.head((out2d, out1d))
end
Flux.@functor Net
Initialize the network instance and transfer it to the appropriate device
model = Net()
device == gpu ? model = gpu(model) : nothing;
Initializing the optimizer with the specified learning rate
optimizer = Flux.Adam(1e-4, (0.9, 0.99));
Let's define functions for training and validation on the same epoch
lossSnet(imgs, features, y) = Flux.Losses.logitcrossentropy(Net(imgs, features), y);
function train_one_epoch(model, Loader, correct_sample, TSamples, Tloss, Optimizer, device)
for (i, (imgs, feat, y)) in enumerate(Loader)
# println("Iteration: ", i)
TSamples += length(y)
imgs = gpu(imgs)
feat = gpu(feat)
y = gpu(y)
wg = Flux.withgradient(Flux.params(model)) do
ŷ = model(imgs, feat)
l = Flux.Losses.logitcrossentropy(ŷ, onehotbatch(y, CLASSES))
return l, ŷ # <- as in your examples: two values
end
batch_loss, y_pred = wg.val
Flux.update!(Optimizer, Flux.params(model), wg.grad)
Tloss += batch_loss
correct_sample += sum(onecold(y_pred, CLASSES) .== y)
end
return Tloss, TSamples, correct_sample
end;
function validate(model, val_loader, task, device, correct_sample, TSamples, running_loss)
preds = nothing
targets = Int[] # an empty Int vector
# n_batches = 0
for (i, (imgs, feat, y)) in enumerate(val_loader)
TSamples += length(y)
imgs = gpu(imgs)
feat = gpu(feat)
y = gpu(y)
ŷ = model(imgs, feat)
running_loss += Flux.Losses.logitcrossentropy(ŷ, onehotbatch(y, CLASSES))
# n_batches += 1
correct_sample += sum(onecold(ŷ, CLASSES) .== y)
end
# avg_loss = running_loss / max(n_batches, 1)
epoch_acc = 100.0 * (correct_sample / TSamples)
epoch_loss = running_loss / TSamples
return epoch_loss, preds, targets, epoch_acc
end
Let's start the network learning cycle
for epoch in 1:10
total_loss = 0.0
train_running_correct = 0
total_samples = 0
correct_sample_val = 0
TSamples_val = 0
running_loss_val = 0
@info "Epoch $epoch"
@info "Workout"
Tloss, TSamples, correct_sample = train_one_epoch(model, train_loader, train_running_correct,
total_samples, total_loss, optimizer, device)
@info "Validation"
avg_loss, preds, targets, epoch_acc_valid = validate(model, val_loader, "val", device, correct_sample_val, TSamples_val, running_loss_val)
epoch_loss = Tloss / TSamples
epoch_acc = 100.0 * (correct_sample / TSamples)
println("loss: $epoch_loss, accuracy: $epoch_acc, val_loss:$avg_loss, val_acc:$epoch_acc_valid")
end
Let's evaluate the model
function evaluate_model_accuracy(loader, model, classes)
total_loss, correct_predictions, total_samples = 0.0, 0, 0
all_preds = []
True_labels = []
for (i, (imgs, feat, y)) in enumerate(loader)
imgs = gpu(imgs)
feat = gpu(feat)
y = gpu(y)
y_pred = model(imgs, feat)
total_loss += Flux.Losses.logitcrossentropy(y_pred, onehotbatch(y, classes))
preds = onecold(y_pred, classes)
true_classes = y
append!(all_preds, preds)
append!(True_labels, true_classes)
correct_predictions += sum(preds .== true_classes)
total_samples += length(y)
end
# Calculating accuracy
accuracy = 100.0 * correct_predictions / total_samples
return accuracy, all_preds, True_labels
end;
Let's evaluate our model by calculating the accuracy on a test dataset.
accuracy_score_LSTM, all_predsLSTMm, true_predS_LSTM = evaluate_model_accuracy(test_loader, model, CLASSES);
println("Accuracy trained model:", accuracy_score_LSTM, "%")
Now let's build an error matrix for visual evaluation of the model.
function plot_confusion_matrix(C)
# Creating a heatmap with large fonts and contrasting colors
heatmap(
C,
title = "Confusion Matrix",
xlabel = "Predicted",
ylabel = "True",
xticks = (1:length(classes), classes),
yticks = (1:length(classes), classes),
c = :viridis, # More contrasting color scheme
colorbar_title = "Count",
size = (600, 400)
)
# Let's add values to the cells for improved visibility.
for i in 1:size(C, 1)
for j in 1:size(C, 2)
annotate!(j, i, text(C[i, j], :white, 12, :bold))
end
end
# We display the graph explicitly
display(current())
end;
preds_for_CM_LSTM = map(x -> x[1], all_predsLSTMm);
conf_matrix = CM.confmat(preds_for_CM_LSTM, true_predS_LSTM)
conf_matrix = CM.matrix(conf_matrix)
plot_confusion_matrix(conf_matrix)
Further improvement of the model implies a better choice of architecture and selection of optimal hyperparameters.
Conclusion
This example shows a workflow for performing radar target classification using machine learning and deep learning techniques. Although this example used synthesized data for training and testing, it can be easily expanded to take into account the actual results of the radar.